Dimensionality reduction beyond neural subspaces
Written on February 26th, 2024 by Heike Stein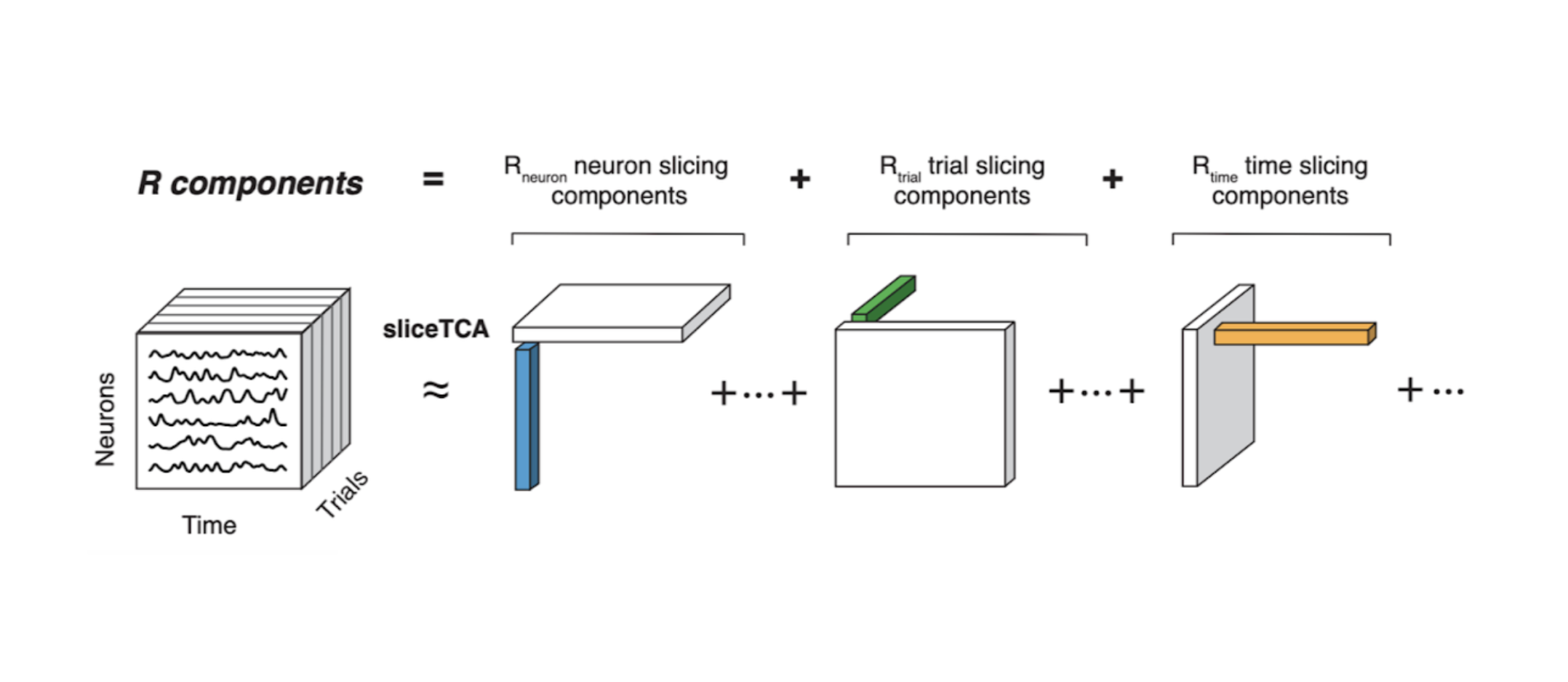
How do we relate activity at the neural population level to behavior? While ever larger recordings have become possible over recent years, it is central to extract low-dimensional structure in this activity in order to test hypotheses about circuit computation in this data. A central tool for this inquiry are dimensionality reduction methods, which identify the most relevant sources of variability in high-dimensional datasets. A basic principle of these methods is the identification of a neural subspace, consisting of a few “latent” dimensions that capture dominant activity patterns, which are often correlated across neurons. The interpretation of low-dimensional neural dynamics is significantly easier than interpreting single-neuron firing patterns in hundreds of neurons, allowing us to hypothesize about single- and multi-area network computations in relation to behavior and to the external world.
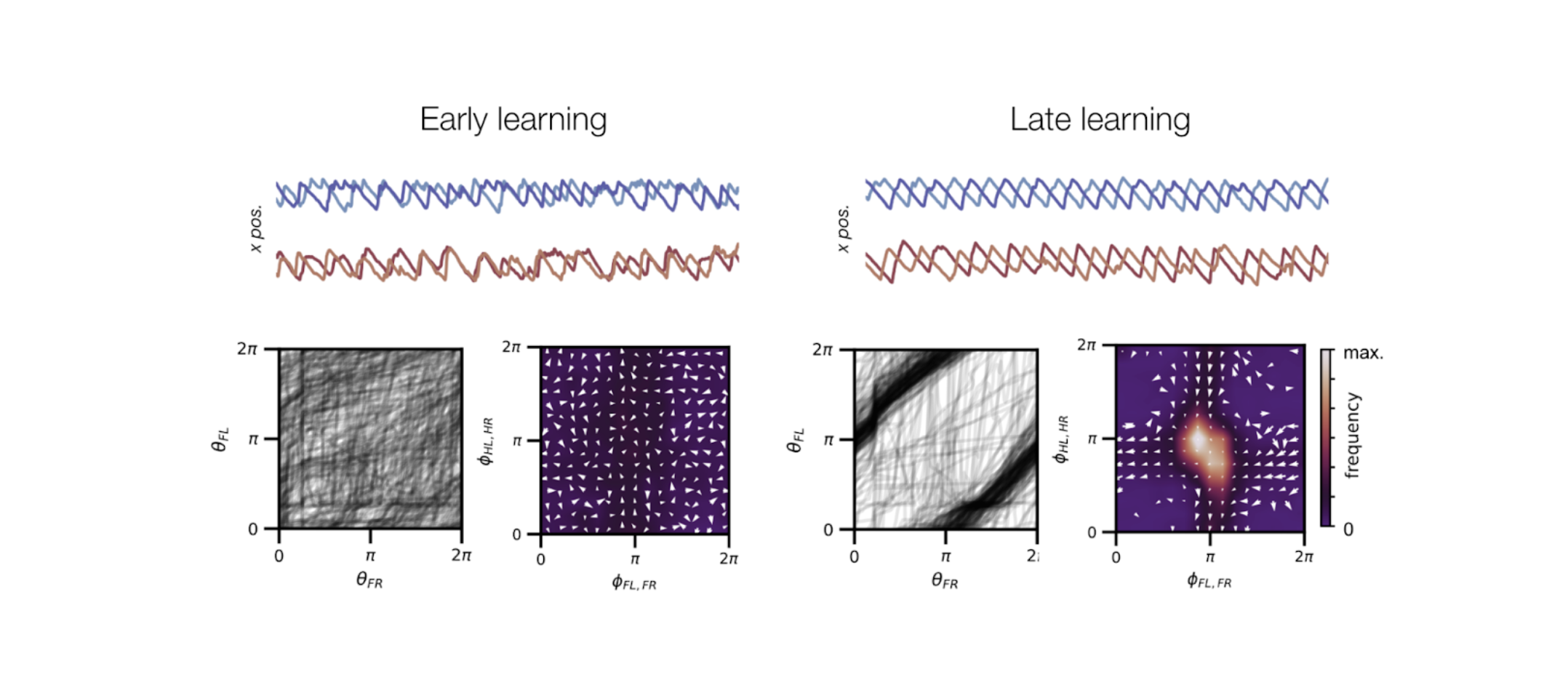
In my postdoc, I studied other relevant classes of covariability in large-scale neural recordings. In particular, in tasks with trial structure, data can be correlated over trials, neurons, or time (rather than just over neurons). Variability in each of these classes can be indicative of the computations performed by a neural population. In this project, we propose a novel, tensor-based dimensionality reduction technique that uncovers different covariability patterns, including neural sequences, trial-to-trial variability in neural firing latencies, and drifting neural subspaces (as is representational drift).
Relevant publications
Pellegrino, A.*, Stein, H.*, Cayco Gajic, N.A. Dimensionality reduction beyond neural subspaces with slice tensor component analysis. Nature Neuroscience (2024).